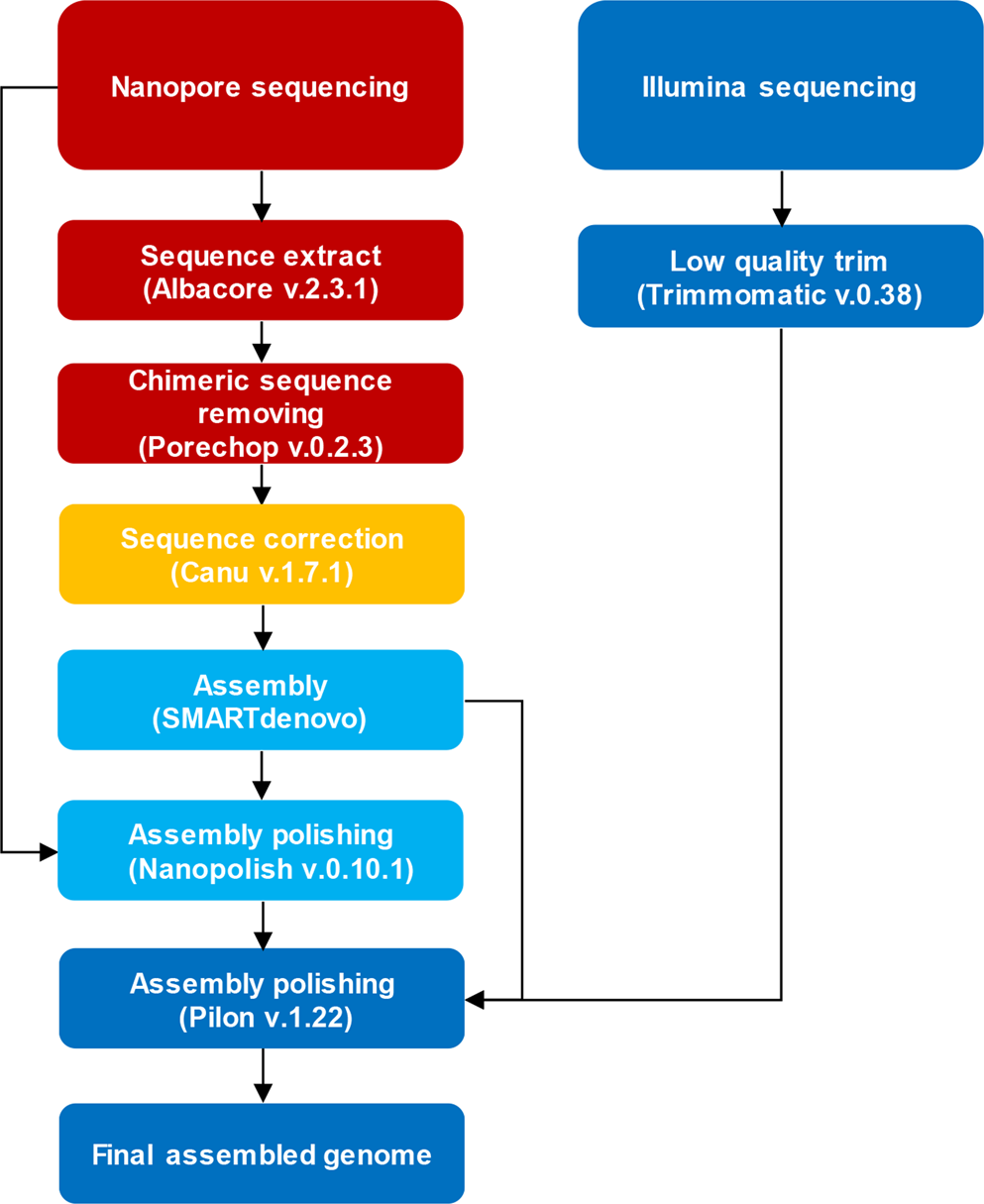
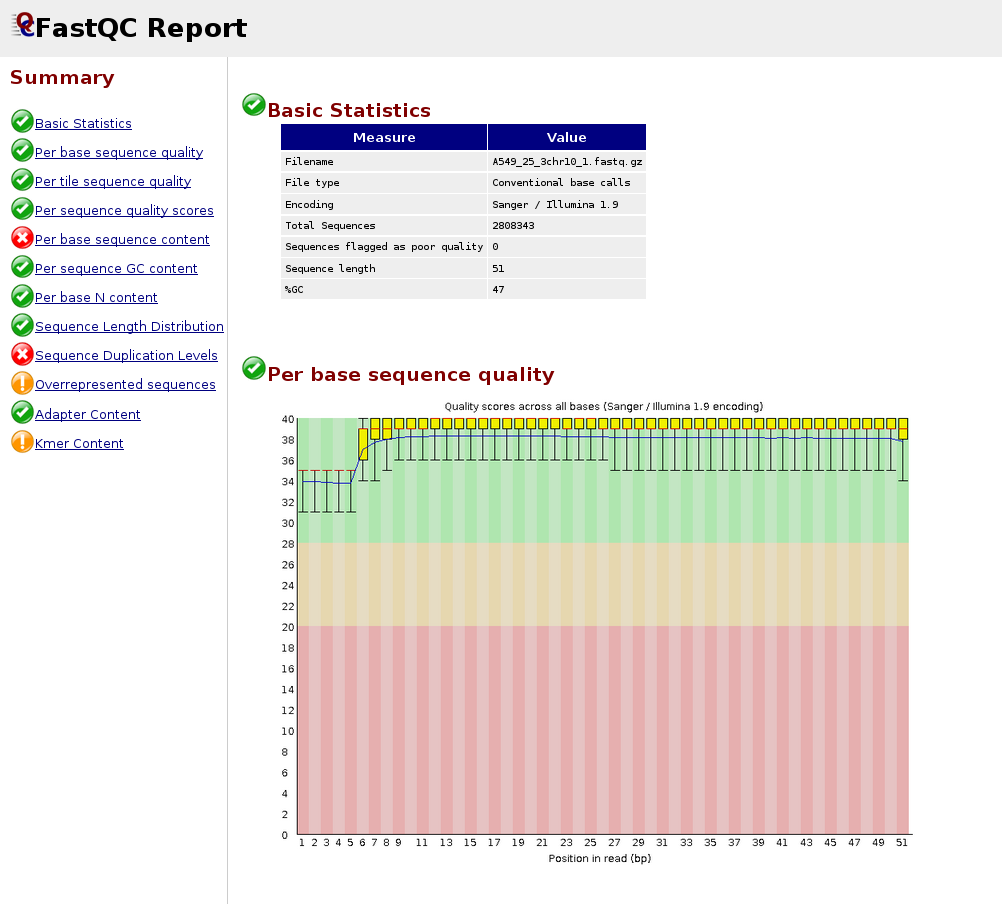
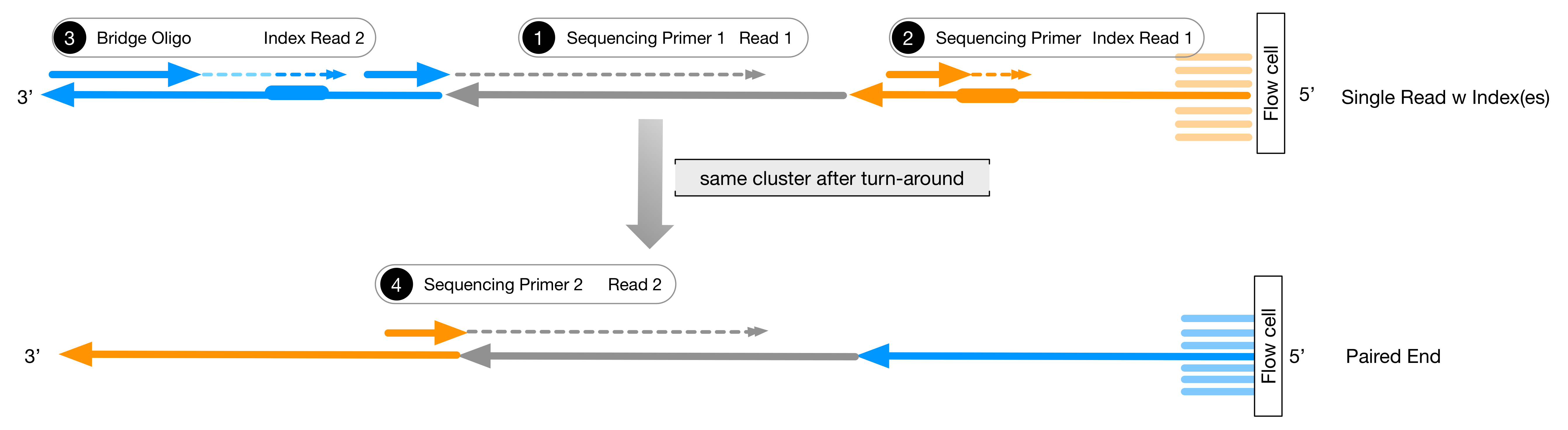
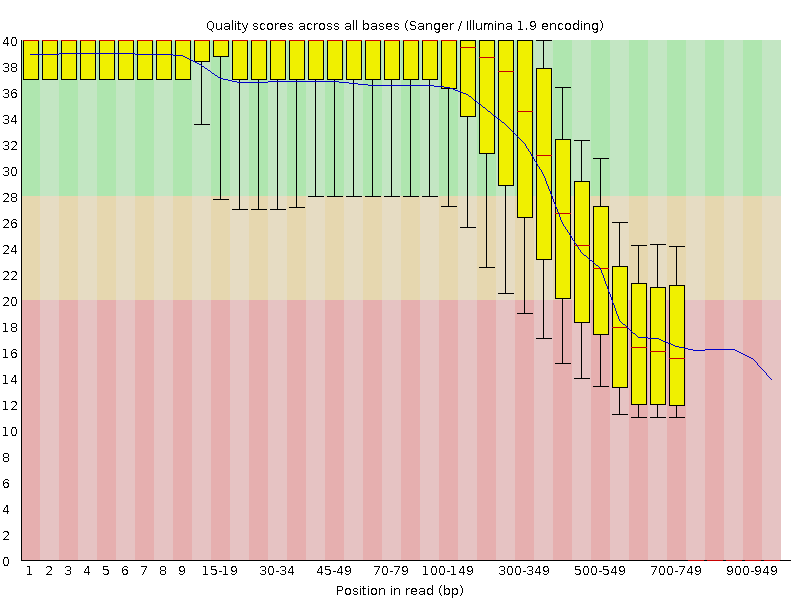
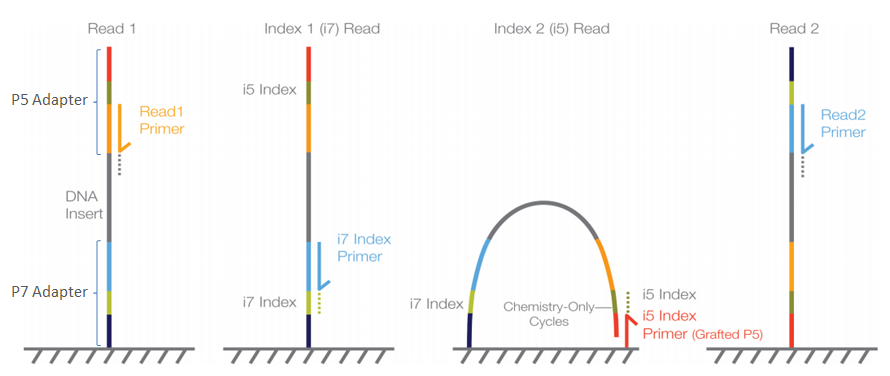
Major short-read and long-read sequencing technologies. (A) Illumina... | Download Scientific Diagram

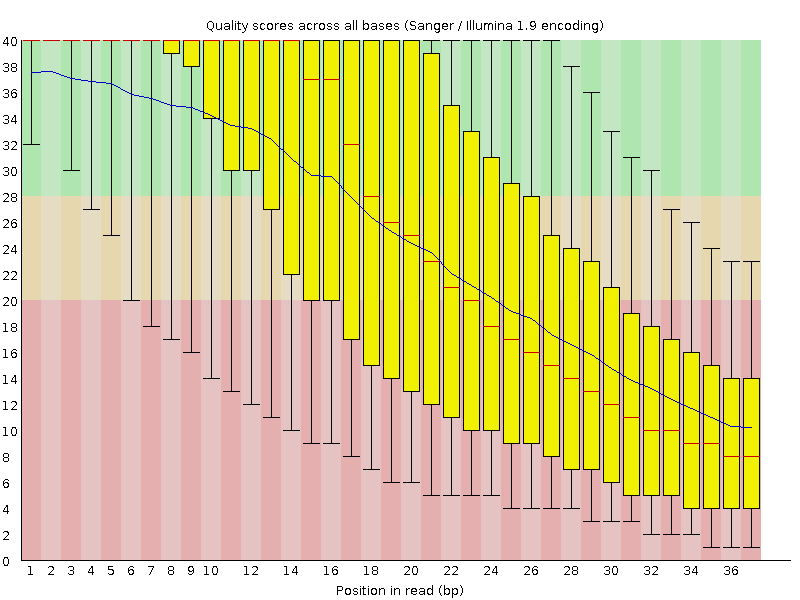
Adapter trimming for single end sRNA-seq data. The reads processed by... | Download Scientific Diagram
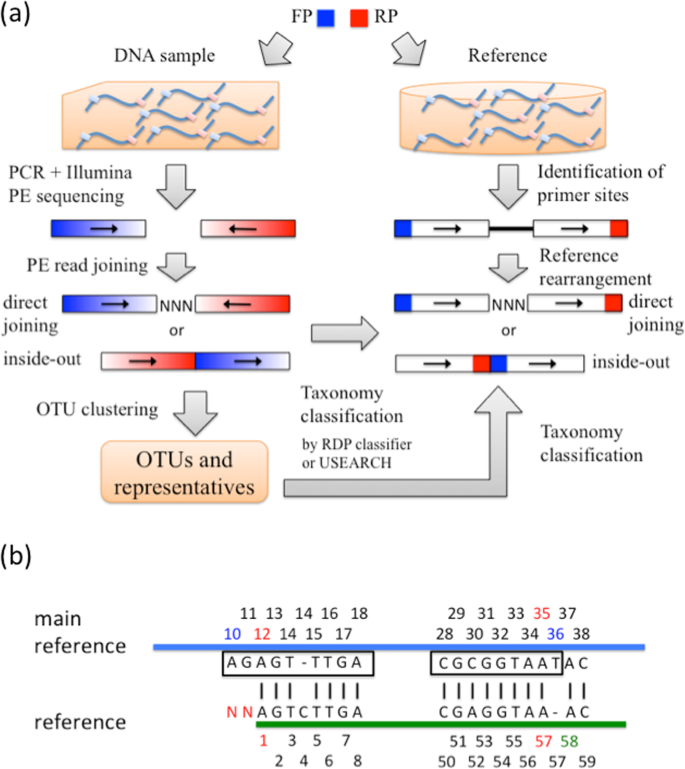
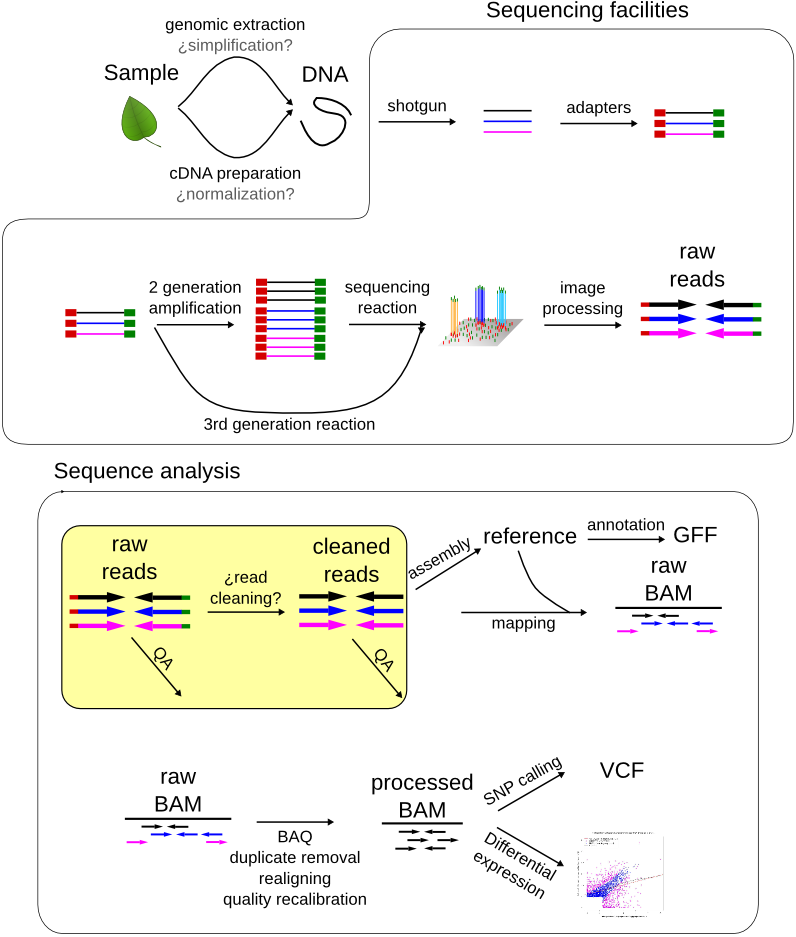
Joining Illumina paired-end reads for classifying phylogenetic marker sequences | BMC Bioinformatics | Full Text

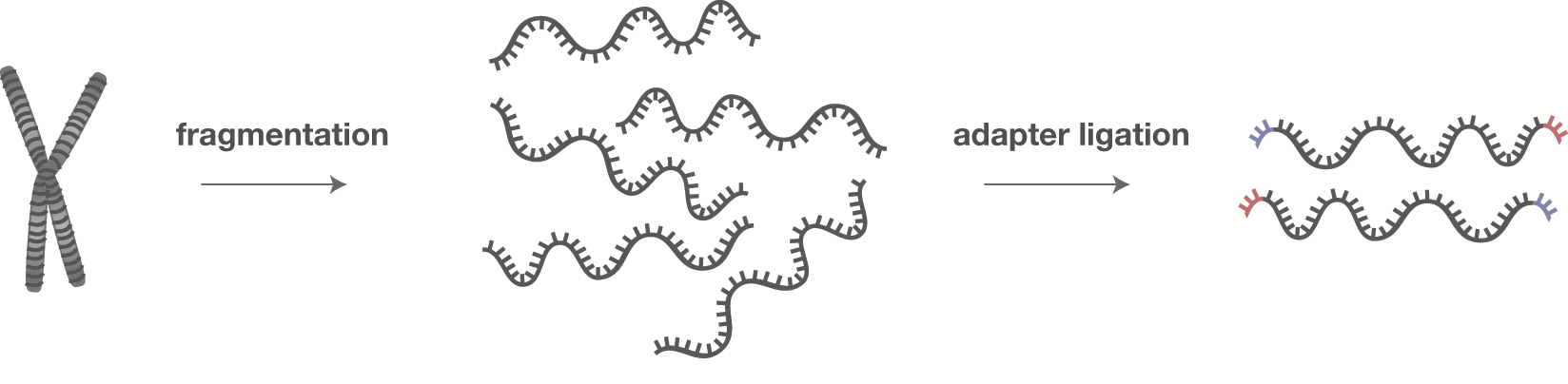
All possible alignments of Cutadapt on 5' end or 3' end with trimming... | Download Scientific Diagram